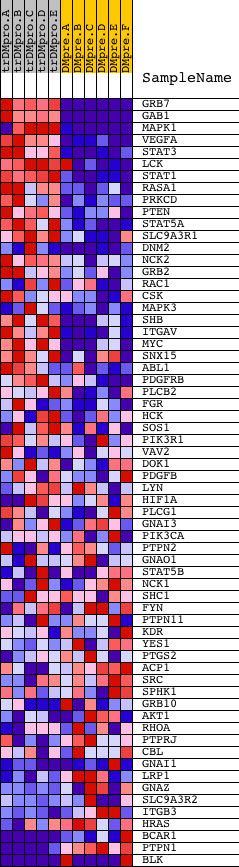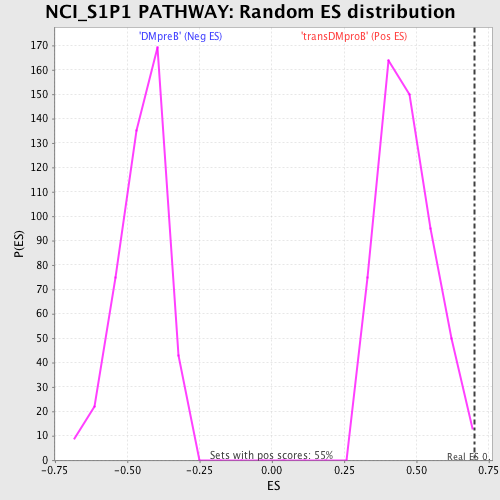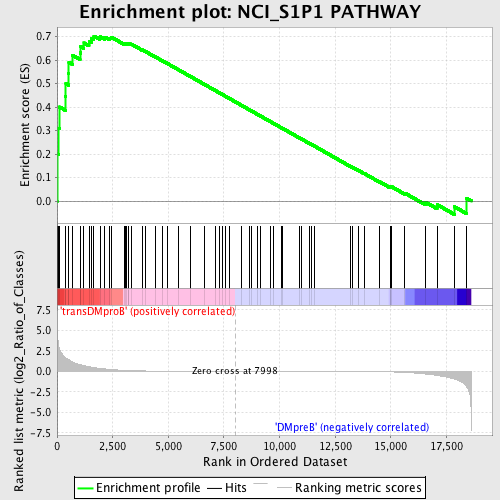
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB.phenotype_transDMproB_versus_DMpreB.cls #transDMproB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_DMpreB.cls#transDMproB_versus_DMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | NCI_S1P1 PATHWAY |
| Enrichment Score (ES) | 0.70235974 |
| Normalized Enrichment Score (NES) | 1.5088001 |
| Nominal p-value | 0.005484461 |
| FDR q-value | 0.2111804 |
| FWER p-Value | 0.975 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GRB7 | 20673 | 5 | 5.748 | 0.2010 | Yes | ||
| 2 | GAB1 | 18828 | 64 | 3.185 | 0.3094 | Yes | ||
| 3 | MAPK1 | 1642 11167 | 108 | 2.667 | 0.4005 | Yes | ||
| 4 | VEGFA | 22969 | 368 | 1.660 | 0.4447 | Yes | ||
| 5 | STAT3 | 5525 9906 | 396 | 1.605 | 0.4994 | Yes | ||
| 6 | LCK | 15746 | 521 | 1.410 | 0.5421 | Yes | ||
| 7 | STAT1 | 3936 5524 | 531 | 1.404 | 0.5908 | Yes | ||
| 8 | RASA1 | 10174 | 698 | 1.106 | 0.6206 | Yes | ||
| 9 | PRKCD | 21897 | 1031 | 0.813 | 0.6312 | Yes | ||
| 10 | PTEN | 5305 | 1062 | 0.791 | 0.6572 | Yes | ||
| 11 | STAT5A | 20664 | 1207 | 0.676 | 0.6732 | Yes | ||
| 12 | SLC9A3R1 | 11250 | 1434 | 0.556 | 0.6805 | Yes | ||
| 13 | DNM2 | 4635 3103 | 1539 | 0.507 | 0.6926 | Yes | ||
| 14 | NCK2 | 9448 | 1655 | 0.455 | 0.7024 | Yes | ||
| 15 | GRB2 | 20149 | 1927 | 0.347 | 0.6999 | No | ||
| 16 | RAC1 | 16302 | 2149 | 0.281 | 0.6978 | No | ||
| 17 | CSK | 8805 | 2369 | 0.230 | 0.6941 | No | ||
| 18 | MAPK3 | 6458 11170 | 2440 | 0.214 | 0.6978 | No | ||
| 19 | SHB | 10493 | 3022 | 0.103 | 0.6701 | No | ||
| 20 | ITGAV | 4931 | 3093 | 0.096 | 0.6697 | No | ||
| 21 | MYC | 22465 9435 | 3120 | 0.093 | 0.6715 | No | ||
| 22 | SNX15 | 23993 3761 | 3200 | 0.085 | 0.6702 | No | ||
| 23 | ABL1 | 2693 4301 2794 | 3222 | 0.082 | 0.6720 | No | ||
| 24 | PDGFRB | 23539 | 3339 | 0.072 | 0.6682 | No | ||
| 25 | PLCB2 | 5262 | 3821 | 0.041 | 0.6438 | No | ||
| 26 | FGR | 4723 | 3971 | 0.034 | 0.6369 | No | ||
| 27 | HCK | 14787 | 4412 | 0.021 | 0.6139 | No | ||
| 28 | SOS1 | 5476 | 4722 | 0.015 | 0.5978 | No | ||
| 29 | PIK3R1 | 3170 | 4940 | 0.013 | 0.5865 | No | ||
| 30 | VAV2 | 5848 2670 | 5455 | 0.008 | 0.5591 | No | ||
| 31 | DOK1 | 17104 1018 1177 | 5995 | 0.006 | 0.5303 | No | ||
| 32 | PDGFB | 22199 2311 | 6629 | 0.003 | 0.4963 | No | ||
| 33 | LYN | 16281 | 7120 | 0.002 | 0.4699 | No | ||
| 34 | HIF1A | 4850 | 7307 | 0.002 | 0.4600 | No | ||
| 35 | PLCG1 | 14753 | 7415 | 0.001 | 0.4543 | No | ||
| 36 | GNAI3 | 15198 | 7576 | 0.001 | 0.4457 | No | ||
| 37 | PIK3CA | 9562 | 7760 | 0.001 | 0.4358 | No | ||
| 38 | PTPN2 | 23414 | 8291 | -0.001 | 0.4073 | No | ||
| 39 | GNAO1 | 4785 3829 | 8644 | -0.002 | 0.3884 | No | ||
| 40 | STAT5B | 20222 | 8729 | -0.002 | 0.3839 | No | ||
| 41 | NCK1 | 9447 5152 | 8993 | -0.002 | 0.3698 | No | ||
| 42 | SHC1 | 9813 9812 5430 | 9137 | -0.003 | 0.3622 | No | ||
| 43 | FYN | 3375 3395 20052 | 9586 | -0.004 | 0.3382 | No | ||
| 44 | PTPN11 | 5326 16391 9660 | 9713 | -0.004 | 0.3315 | No | ||
| 45 | KDR | 16509 | 10086 | -0.005 | 0.3117 | No | ||
| 46 | YES1 | 5930 | 10143 | -0.005 | 0.3088 | No | ||
| 47 | PTGS2 | 5317 9655 | 10875 | -0.008 | 0.2697 | No | ||
| 48 | ACP1 | 4317 | 10964 | -0.008 | 0.2652 | No | ||
| 49 | SRC | 5507 | 11341 | -0.009 | 0.2453 | No | ||
| 50 | SPHK1 | 5484 1266 1402 | 11415 | -0.010 | 0.2417 | No | ||
| 51 | GRB10 | 4799 | 11584 | -0.010 | 0.2330 | No | ||
| 52 | AKT1 | 8568 | 13176 | -0.023 | 0.1481 | No | ||
| 53 | RHOA | 8624 4409 4410 | 13271 | -0.024 | 0.1438 | No | ||
| 54 | PTPRJ | 9664 | 13565 | -0.028 | 0.1290 | No | ||
| 55 | CBL | 19154 | 13815 | -0.033 | 0.1168 | No | ||
| 56 | GNAI1 | 9024 | 14511 | -0.051 | 0.0811 | No | ||
| 57 | LRP1 | 9284 | 14976 | -0.073 | 0.0587 | No | ||
| 58 | GNAZ | 71 | 14990 | -0.074 | 0.0605 | No | ||
| 59 | SLC9A3R2 | 23089 1544 1514 | 15037 | -0.077 | 0.0608 | No | ||
| 60 | ITGB3 | 20631 | 15630 | -0.137 | 0.0337 | No | ||
| 61 | HRAS | 4868 | 16569 | -0.329 | -0.0054 | No | ||
| 62 | BCAR1 | 18741 | 17079 | -0.500 | -0.0153 | No | ||
| 63 | PTPN1 | 5325 | 17861 | -0.942 | -0.0244 | No | ||
| 64 | BLK | 3205 21791 | 18413 | -1.858 | 0.0109 | No |